Neuropharmacology
Neuropharmacology is the study of how drugs affect the molecular, cellular or behavioral functions of the central and peripheral nervous system. Faculty in our department use modern tools of neuroscience to investigate fundamental brain processes implicated in several neurological disorders — including epilepsy, addiction, ataxia, dystonia, intellectual disability, Parkinson's disease, neuropathic pain and Alzheimer's disease — then translate this knowledge into discovery and development of novel therapeutic strategies.
Our Work
Examining How Genetics Can Influence Epilepsy in Children
Jennifer Kearney, PhD, associate professor of Pharmacology, works in conjunction with the clinicians at Northwestern Memorial Hospital and the Ann and Robert H. Lurie Children’s Hospital of Chicago to learn how an individual’s genetic background influences epilepsy.
Neuronal Signaling at a Molecular and Cellular Level
Geoff Swanson, PhD, professor of Pharmacology, studies how brain cells communicate with each other during normal circumstances, during memory processes and how the processes go wrong in disease.
Developing Mechanistic Based Therapeutic Opportunities for Parkinson’s Disease
Loukia Parisiadou, PhD, assistant professor of Pharmacology, uses a multidisciplinary approach spanning cellular, molecular, network and behavioral levels to understand the molecular basis of Parkinson’s disease.
Pioneering Academic Drug Discovery
Daniel Martin Watterson, PhD, professor of pharmacology and the John G. Searle Professor of Molecular Biology and Biochemistry, studies biological mechanisms important in how cells communicate with each other. The work is advancing basic and translational knowledge about critical biological processes and molecules that regulate physiological pathways, and how they are altered in diseases such as Alzheimer’s disease, brain injury and cancer. The goal is to develop novel drug treatments that can intervene in disease progression.
Investigating Why Mutations in Ion Channels Cause Disease
Paul DeCaen, PhD, associate professor of Pharmacology, studies why a class of proteins called ion channels cause diseases. His goal is to intervene with these diseases and either keep them from happening, or perhaps, control them after they've already manifested.
Investigating the molecular and cellular physiology of calcium signaling
Murali Prakriya, PhD, the Magerstadt Professor of Pharmacology and professor of Medicine (Allergy and Immunology), studies how cellular calcium signals are generated and how these calcium signals impact the functional behavior of neurons, astrocytes, and immune cells.
Neuropharmacology Research Labs
Shana Augustin LabStudying the molecular mechanisms of synaptic transmission and modulation by GPRCs to control motor actions and decision making
Research Description
Using an integrative approach – combining physiology, behavior and optical imaging – the Augustin lab studies the molecular mechanisms of synaptic transmission, and its modulation by GPRCs to control motor actions and decision making. We are particularly interested in the flow of information through the cortico-basal ganglia circuits and how this is disrupted with alcohol and substance use disorder. Our lab uses an integrative approach combining physiology, behavior, and optical imaging techniques.
For more information, see Dr. Augustin's faculty profile or the Augustin Lab website.
Publications
See Dr. Augustin's publications on PubMed.
Contact
Contact Dr. Augustin.
Graduate Student: Yasmin Lowe
Post-Baccalaureate Research Fellow: Anthony James, Jr.
Paul DeCaen LabStudying ion channel relevance in cell biology and disease progression
Studying ion channel relevance in cell biology and disease progression
Research Description
We study the biophysics, pharmacology and physiology of ion channels. Currently, we are focused on two divergent groups: voltage gated sodium channels (Nav) and Polycystin channels (also called Polycystic Kidney Disease Proteins, PKDs). Aside from these foci, we actively explore novel ion channels found in prokaryotic and eukaryotic cells with the goal of understanding their function in cell physiology.
Current Projects
Voltage Gated Sodium Channels
Navs conduct sodium ions into excitable cells like muscle and neurons, causing the cell membrane to depolarize on the microsecond time scale, a process essential for rapid communication in multicellular organisms. Potentially fatal conditions such as forms of epilepsy and cardiac arrhythmias arise when Navs are mutated.
With our collaborators, we continue to examine key questions:
- How do these transmembrane proteins sense electrical potential and change from nonconductive to conductive states?
- How do these transmembrane proteins select for sodium ions and not allow passage of the other ions present?
- What are the mechanisms of action of clinically relevant drugs (e.g. Valproate and Lamotrigine) and where are their receptor sites?
Polycystin Channels and Primary Cilia
Mutations in PKD1 and PKD2 are associated with Autosomal Dominant Kidney Disease (ADPKD). ADPKD is one of the most common monogenetic diseases in mankind, where progressive cyst formation results in kidney failure. Several members of the polycystins (PKD1, PKD1-L1, PKD2 and PKD2-L1) have been found in the primary cilia from cells of various tissues besides the kidney. The primary cilium is a solitary, small (5-15 mM in length) protuberance from the apical side of polarized cells.
With help from our collaborators, our research is directed to answer key questions:
- How do ADPKD mutations alter PKD2 function? Do some mutations ‘turned on’ while others ‘turn off ’ the PKD2 channel?
- How does PKD1/2 channel dysfunction result in cyst formation? Or conversely, what normal function do they serve for the primary cilium and how do PKDs maintain cell polarity?
- What are the receptor sites within PKD2s that can modulation its ion channel function and are they drug-able?
For lab information and more, see Dr. DeCaen's faculty profile and lab website.
Publications
See Dr. DeCaen's publications on PubMed.
Contact
Contact Dr. DeCaen at 312-503-5930.
Postdoctoral Fellows: Orhi Esarte Palomero, Louise Vieira
Graduate Students: Eduardo Guadarrama, Megan Larmore
Research Staff: Victoria Pappas
Al George LabInvestigating the structure, function, pharmacology and molecular genetics of ion channels and channelopathies
Investigating the structure, function, pharmacology and molecular genetics of ion channels and channelopathies
Research Description

Ion channels are ubiquitous membrane proteins that serve a variety of important physiological functions, provide targets for many types of pharmacological agents and are encoded by genes that can be the basis for inherited diseases affecting the heart, skeletal muscle and nervous system.
Dr. George's research program is focused on the structure, function, pharmacology and molecular genetics of ion channels. He is an internationally recognized leader in the field of channelopathies based on his important discoveries on inherited muscle disorders (periodic paralysis, myotonia), inherited cardiac arrhythmias (congenital long-QT syndrome) and genetic epilepsies. Dr. George’s laboratory was first to determine the functional consequences of a human cardiac sodium channel mutation associated with an inherited cardiac arrhythmia. His group has elucidated the functional and molecular consequences of several brain sodium channel mutations that cause various familial epilepsies and an inherited form of migraine. These finding have motivated pharmacological studies designed to find compounds that suppress aberrant functional behaviors caused by mutations.
Recent Findings
- Discovery of novel, de novo mutations in human calmodulin genes responsible for early onset, life threatening cardiac arrhythmias in infants and elucidation of the biochemical and physiological consequences of the mutations.
- Demonstration that a novel sodium channel blocker capable of preferential inhibition of persistent sodium current has potent antiepileptic effects.
- Elucidation of the biophysical mechanism responsible for G-protein activation of a human voltage-gated sodium channel (NaV1.9) involved in pain perception.
Current Projects
- Investigating the functional and physiological consequences of human voltage-gated sodium channel mutations responsible for either congenital cardiac arrhythmias or epilepsy.
- Evaluating the efficacy and pharmacology of novel sodium channel blockers in mouse models of human genetic epilepsies.
- Implementing high throughput technologies for studying genetic variability in drug metabolism.
- Implementing automated electrophysiology as a screening platform for ion channels.
For lab information and more, see Dr. George’s faculty profile.
Publications
See Dr. George's publications on PubMed.
Contact
Contact Dr. George at 312-503-4892.
Research Faculty: Thomas Lukas, Christopher Thompson, Carlos Vanoye
Faculty Contributors: Brandon Chalazan
Senior Researchers: Reshma Desai, Jean-Marc Dekeyser, Christine Simmons
Lab Manager: Tatiana Abramova
Postdoctoral Fellows: Dina Simkin
Medical Residents: Scott Adney, Tracy Gertler
Graduate Students: Surobhi Ganguly, Dalton Huey Adil Wafa, Lisa Wren
Technical Staff: Alexandra Hong
Jennifer Kearney LabInvestigating the genetic basis of epilepsy
Investigating the genetic basis of epilepsy
Research Description

My research program is focused on studying the genetic basis of epilepsy, a common neurological disorder that affects approximately 1% of the population. Epilepsy is thought to have a genetic basis in approximately two-thirds of patients, including a small fraction caused by single gene mutations. Many genes responsible for rare, monogenic epilepsy have been identified. The genes identified are components of neuronal signaling, including voltage-gated ion channels, neurotransmitter receptors, ion-channel associated proteins and synaptic proteins. We use mouse models with mutations in ion channel genes to understand the underlying molecular basis of epilepsy and to identify modifier genes that influence phenotype severity by modifying disease risk. Identifying genes that influence epilepsy risk improves our understanding of the underlying pathophysiology and suggests novel targets for therapeutic intervention.
For lab information and more, see Dr. Kearney's faculty profile or lab site.
Publications
See Dr. Kearney's publications on PubMed.
Contact
Contact Dr. Kearney at 312-503-4894.
Research Faculty: Nicole Hawkins
Graduate Student: Emilia Gibes
Technical Staff: Levi Barse, Nathan Speakes, Tyler Thenstedt
Richard Miller LabStudying molecular aspects of nerve cell communication and neurodegenerative disease
Studying molecular aspects of nerve cell communication and neurodegenerative disease
Research Description
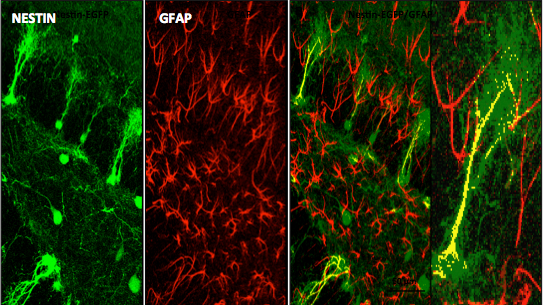
The laboratory led by Richard Miller, PhD, is interested in studying molecular aspects of nerve cell communication. One of our major interests has been to understand the structure and function of calcium channels. The influx of Ca into neurons through these channels is important for many reasons, including the release of neurotransmitters. We have identified a family of molecules that act as Ca channels in neurons and other types of cells. Each of these molecules has slightly different properties that underlie different neuronal functions. We have analyzed the properties of these molecules by examining their electrophysiological properties following their expression in heterologous expression systems and imaging techniques. Furthermore, we have generated calcium channel knockout mice that have interesting properties such as altered pain thresholds, seizures and memory deficits. We have also been interested in how Ca channels can be regulated by the activation of Gprotein coupled receptors. We have been analyzing the interaction of Gprotein subunits with Ca channels using FRET imaging and other techniques.
Other projects in our laboratory are aim to understanding the molecular basis of neurodegenerative disease. We study Alzheimer's disease, Amyotrophic lateral sclerosis (Lou Gehrig's disease), HIV-1 related dementia and other neuropathological conditions. In the case of HIV-1 infection, we have been examining the properties and functions of HIV-1 receptors on neurons. These receptors are known to be receptors for chemokines -small proteins that are known to direct the functions of the immune system. We have shown that neurons express many types of chemokine receptors and that activation of these receptors can produce both short and long term effects on neurons. Activation of chemokine receptors expressed by sensory neurons produces neuronal excitation and pain. Activation of chemokine receptors on hippocampal neurons has a prosurvival effect, whereas binding of HIV-1 to these receptors induces apoptosis. We are studying the molecular mechanisms that produce this diverse effects with a view to understanding the molecular basis for HIV-1 related dementias.
For lab information and more, see Dr. Miller’s faculty profile.
See Dr. Miller’s blog "The Keys to all Mythologies: Science, Medicine and Magic" to read articles concerning scientific topics of current interest as well as historical accounts of scientific issues.
Publications
See Dr. Miller's publications on PubMed.
Contact
Contact Dr. Miller at 312-503-3211.
Research Associate: Dongjun Ren
Loukia Parisiadou LabInvestigating the cellular and molecular pathways by which mutations in genes linked to Parkinson’s Disease contribute to disease pathogenesis
Investigating the cellular and molecular pathways by which mutations in genes linked to Parkinson’s Disease contribute to disease pathogenesis
Research Description
Parkinson’s disease (PD) has been classically considered a sporadic disease, however, it is now recognized to have a substantial genetic component. Interestingly, the same genes involved in the autosomal-dominantly inherited forms of PD such as SNCA and LRRK2 can act as risk factors in idiopathic cases of PD, as well. Therefore, studying the pathophysiological functions of these PD-related genes could provide valuable insights into the process of understanding the underlying pathogenic mechanisms of PD.
A large part of Dr. Parisiadou’s scientific efforts was focused on functionally characterizing the LRRK2 protein, and a number of relevant studies provided insights on the undetermined role of LRRK2 protein, as well as revealed an important functional interplay between SNCA and LRRK2. Based on the previous knowledge, Loukia Parisiadou’s research program is focused on the delineation of the contribution of LRRK2 and SNCA mutations in PD. To achieve this, a variety of mouse genetic, neuronal culture, histology, cell biology, biochemistry, and behavioral approaches will be utilized. Given the relevance of LRRK2 and α-synuclein with the sporadic forms of PD, our long-term research goal by interrogating α-synuclein and mainly LRRK2-dependent alterations at the cellular, network and behavioral levels is to appreciate the pathophysiology of PD. While there is still a long way to go in understanding the etiology of PD, LRRK2, and SNCA mutations have provided important insights into this process, and it is expected to have a crucial role to this effort for the following years to come.
Recent Findings
- LRRK2 directs neurite morphology by the regulation of cytoskeletal dynamics in neurons
- A functional interplay between alpha-synuclein and LRRK2, two gene products linked to autosomal-dominantly inherited forms of PD.
- An emerging function of LRRK2 at the synapse: LRRK2 directs synaptogenesis, and neurotransmission.
Current Projects
- LRRK2-mediated deregulation of PKA activity results in aberrant dopaminergic and corticostriatal signaling in the striatal projection neurons.
- Looking for novel therapeutic approaches for LRRK2 besides kinase inhibition: the identification of novel LRRK2 modifiers in specific sub-neuronal populations.
- Unraveling the mechanistic details of α-synculein clearance.
For lab information and more, see Dr. Parisiadou's faculty profile and lab website and watch the video, "Developing Mechanistic Based Therapeutic Opportunities for Parkinson’s Disease."
Publications
See Dr. Parisiadou's publications on PubMed.
Contact
Contact Dr. Parisiadou at 312-503-2652.
Postdoctoral Fellows: Chuyu Chen, Giulia Tombesi
Graduate Student: Ciarra Smith
Research Technician: Vanessa Promes
Murali Prakriya LabCalcium signaling, inflammation, and brain function
Calcium signaling, inflammation, and brain function
Research Description
Research in the laboratory of Murali Prakriya, PhD, is focused on the molecular and cellular mechanisms of intracellular calcium (Ca2+) signaling. Ca2+ is one of the most ubiquitous intracellular signaling messengers, mediating many essential functions including gene expression, chemotaxis and neurotransmitter release. Cellular Ca2+ signals generally arise from the opening of Ca2+-permeable ion channels, a diverse family of membrane proteins. We are studying Ca2+ signals arising from the opening of store-operated Ca2+ channels (SOCs). SOCs are found in the plasma membranes of virtually all mammalian cells and are activated through a decrease in the calcium concentration ([Ca2+]) in the endoplasmic reticulum (ER), a vast membranous network within the cell that serves as a reservoir for stored calcium. SOC activity is stimulated by a variety of signals such as hormones, neurotransmitters and growth factors whose binding to receptors generates IP3 to cause ER Ca2+ store depletion.
The best-studied SOC is a sub-type known as the Ca2+ release activated Ca2+ (CRAC) channel encoded by the Orai1 protein. CRAC channels are widely expressed in immune cells and generate Ca2+ signals important for gene expression, proliferation and the secretion of inflammatory mediators. Loss of CRAC channel function due to mutations in CRAC channel genes leads to a devastating immunodeficiency syndrome in humans. Our goals are to understand the molecular mechanisms of CRAC channel activation, and their physiological roles especially in the microglia and astrocytes of the brain, and in the airway epithelial cells of the lung.
Recent Findings
Despite the fact that CRAC channels are found in practically all cells, their properties and functions outside the immune system remain largely unexplored. In order to fill this gap, we have begun investigation of CRAC channel properties and their functions in two major organ systems: in the brain and the lung.
- In the brain, we are studying the role of CRAC channels for dendritic Ca2+ signaling in excitatory neurons of the hippocampus, and their role in synaptic plasticity and cognitive functions. We have found that CRAC channels formed by Orai1 are critical for amplifying glutamate receptor evoked calcium signals in dendritic spines of hippocampal neurons, and this step is essential for driving structural and functional measures of synaptic plasticity and cognitive processes involving learning and memory.
- In a second project, we are studying the role of CRAC channels in driving neuroinflammation. We have found that CRAC channels formed by Orai1 are essential for the production and release of proinflammatory cytokines and chemokines in microglia and astrocytes. We are examining the relevance of this pathway for mediating inflammatory and neuropathic pain.
- A third project is examining the role of CRAC channels for mediating pro- and anti-inflammatory processes in the lung. We have found that CRAC channels are a major mechanism for mobilizing Ca2+ signals in lung epithelial cells, and the downstream production of both pro- and anti-inflammatory mediators. We are examining the relevance of this signaling for lung inflammation in the context of asthma.
For lab information and more, see Dr. Prakriya’s faculty profile and lab website.
Publications
See Dr. Prakriya's publications on PubMed.
Contact
Contact Dr. Prakriya at 312-503-7030.
Research Faculty: Megumi Yamashita
Postdoctoral Fellows: Kirill Korshunov, Aashutosh Shetti
Graduate Students: Kaitlyn DeMeulenaere, Se’ Ferrell, Michaela Novakovic
Technical Staff: Megan Martin
Richard Smith LabIon channel contributions to brain development and disease
Ion channel contributions to brain development and disease
Research Description
Dr. Smith’s research examines ion channel contributions to human brain development and brain malformation diseases. Since 2016 Dr. Smith has received continuous funding support from the National Institute of Health to evaluate the cellular, biophysical, and genetic mechanisms underlying aberrant neurophysiology related to developmental channelopathies.
Current Projects
Ion channel diseases affecting brain development.
Evaluating patients with cerebral cortex malformation diseases, we identified a role for a sodium channel (SCN3A), and a Na+/K+-ATPase (ATP1A3) in the developing human cortex. This developmental channelopathy affects several populations of cells in the fetal human cortex, which at critical times of development results in atypical neocortical formation.
Bioelectricity in human brain development and evolution.
We use animal model systems (i.e. mouse and ferret) to unravel the non-canonical roles of ion channels in generating “Bioelectric” patterns of neuronal activity across mammalian brain development. In conjunction with Human Genetics studies, these studies enable dissection of species-specific neurophysiology, identifying key cellular and biophysical mechanisms unique to the human brain and disease pathology.
Human induced pluripotent stem cell (iPSCs) screening and neuronal disease modeling platform.
Using human iPSCs, we model various ion channel diseases found in human patients. In these neuronal culture systems, we use both patch-clamp and high-throughput neurophysiology assays to evaluate pharmacology and precision therapeutics.
For more information, see Dr. Smith's faculty profile or lab site.
Publications
See Dr. Smith's publications on PubMed.
Contact
Contact Dr. Smith.
Graduate Student: Alex Briegel
Undergraduate Student: Pranav Inampudi
Technical Staff: Madeline Burke, Sean Golinski, Karla Soriano
Geoffrey Swanson LabStudying glutamate receptors in the modulation of neurotransmission and induction of synaptic plasticity
Studying glutamate receptors in the modulation of neurotransmission and induction of synaptic plasticity
Research Description
Geoffrey Swanson’s, PhD, laboratory studies the molecular and physiological properties of receptor proteins that underlie excitatory synaptic transmission in the mammalian brain. Current research focuses primarily on understanding the roles of kainate receptors, a family of glutamate receptors whose diverse physiological functions include modulation of neurotransmission and induction of synaptic plasticity. We are also interested in exploring how kainate receptors might contribute to pathological processes such as epilepsy and pain. The laboratory investigates kainate receptor function using a diverse group of techniques that include patch-clamp electrophysiology, selective pharmacological compounds, molecular and cellular techniques and gene-targeted mice.
Current Projects
- Isolation and characterization of new marine-derived compounds that target glutamate receptors
- Kainate receptors in hippocampal synaptic transmission
- Mechanisms of kainate receptor assembly and trafficking
For lab information and more, see Dr. Swanson’s faculty profile and lab website.
Publications
See Dr. Swanson's publications on PubMed.
Contact
Contact Dr. Swanson at 312-503-1052.
Graduate Student: Brynna Webb
Technical Staff: Helene Lyons-Swanson
Martin Watterson LabFocusing on the role of protein phosphorylation pathways in disease onset and progression and their potential as drug discovery targets
Focusing on the role of protein phosphorylation pathways in disease onset and progression and their potential as drug discovery targets
Research Description
Current Projects
The role of calmodulin (CaM) mediated signal transduction pathways in physiology and pathophysiology
-
Using of emerging technologies to understand how CaM and a CaM-regulated enzyme could be encoded, expressed, regulated and assembled into a calcium signal transduction complex
-
Using of integrative (in vivo) chemical biology and molecular genetics to gain insight into how landmark CaM-regulated protein kinases are involved in physiology and pathophysiology
Integrative chemical biology and development of novel therapeutics for attenuation of disease progression
-
Using the “smart chemistry” approach integrated with “smart biology” screens for rapid discovery of novel small molecules with potential use in targeting pathophysiology progression related to diseases ranging from neurological disorders, cancer, inflammatory conditions, cardiovascular and pulmonary disease
-
Discovering and developing novel small molecule compounds that selectively attenuate the increased production of proteins called proinflammatory cytokines, which can cause tissue injury and disease when produced in excess
We ultimately hope to find, by targeting pathophysiology mechanisms which contribute to disease progression, a series of novel small molecules with potential to be effective against a variety of disorders.
For lab information and more, see Dr. Watterson’s faculty profile.
Publications
Contact
Contact Dr. Watterson at 312-503-0657.
Adjunct Associate Professor: Jeff Pelletier
Research Associate Professor: Luda Shuvalov
Research Assistant Professor: Saktimayee Roy
Ziarek LabExploring the protein structure/function/dynamics relationship, molecular mechanisms of allostery, and the rational design of molecular therapeutics and tools – with a long-standing interest in G protein-coupled receptors (GPCRs)
Exploring the protein structure/function/dynamics relationship, molecular mechanisms of allostery, and the rational design of molecular therapeutics and tools – with a long-standing interest in G protein-coupled receptors (GPCRs)
Research Description
G protein-coupled receptors (GPCRs) dominate the therapeutic market – being the targets for more than 30 percent of FDA-approved drugs. They also recognize most drugs of abuse including opioids, cannabinoids, LSD, cocaine, and methamphetamine. Unlike many signaling proteins that function as binary switches between 'on and off' states, GPCRs feature a ligand-independent basal activity that is increased or decreased upon orthosteric ligand binding, and then further regulated by allosteric modulators. A single receptor may specifically recognize several ligands and respond uniquely to each, creating a complex conformational landscape. There is immense therapeutic potential in the ability to tune receptor signaling using partial agonists, biased agonists or allosteric modulators – and it remains poorly understood and virtually untapped.
The long-term vision of the Ziarek Lab is to decipher the molecular mechanisms of GPCR allostery for rational drug discovery. The lab's current understanding of biased agonism derives primarily from cell-based functional assays that are easily complicated by receptor expression levels, off-target effects and cell type. Integrating biophysical studies of purified protein with this cellular information would permit a complete molecular understanding. Some research areas the Ziarek lab is currently making progress on are:
- investigating the conformational space of wildtype GPCRs using a combined biophysical and computational approach;
- characterizing the allosteric mechanisms of arrestin activation;
- developing novel 19F-NMR labels, pulse programs and NMR data analysis pipelines for expanding the molecular weight limits of NMR;
- testing if arrestin’s local disorder and conformational entropy play a role in receptor and scaffolding function; and
- actualizing the full potential of NMR to explore GPCR conformational dynamics using receptors compatible with E. coli expression systems.
For more information, see Dr. Ziarek's faculty profile and laboratory website.
Bumbak, F., Bower, J.B., Zemmer, S.C., Pons, M., Inoue, A., Paniagua, J.C., Yan, F., Ford, J., Wu, H., Robson, S.A., Bathgate, R.A.D., Scott, D.J., Gooley, P.R. & Ziarek, J.J. 2023. Stabilization of pre-existing neurotensin receptor conformational states by arrestin-1 and the biased allosteric modulator ML314. Nature Communications, 14(1):3328. 10.1038/s41467-023-38894-8.
- We used 13CεH3-methionine NMR spectroscopy to show that binding of phosphatidylinositol-4,5-bisphosphate (PIP2) to the receptor’s intracellular surface allosterically tunes the timescale of motions at the orthosteric pocket and conserved activation motifs – without dramatically altering the structural ensemble. A β-arrestin biased allosteric modulator transforms the NTS1:G protein complex into a concatenation of substates, without triggering transducer dissociation, suggesting that it may function by stabilizing signaling incompetent G protein conformations such as the non-canonical state. Together, this work demonstrates the importance of kinetic information to a complete picture of the GPCR activation landscape.
Bumbak, F., Pons, M., Inoue, A., Paniagua, J.C., Yan, F., Wu, H., Robson, S.A., Bathgate, R.A.D., Scott, D.J., Gooley, P.R. & Ziarek, J.J. 2023. Ligands tune the local and global motions of neurotensin receptor 1 (NTS1). Cell Reports, 42(1): 112015. https://doi.org/10.1016/j.celrep.2023.112015.
- Here, we expanded a novel method comparing experimental NMR data to density functional theory (DFT) quantum calculations. Our results suggest a role for sub-microsecond dynamics and conformational entropy in GPCR ligand discrimination. This approach sidesteps the isotopic labelling limitations imposed by eukaryotic expression systems. Future studies aim to test the generalizability of our results to the GPCR superfamily and quantitate the underlying motions.
- In this work, we introduce cysteine-reactive 19F probes at the intracellular tip of NTS1 transmembrane helix 6 (TM6). We observe a linear three-site exchange process that is both thermodynamically and kinetically remodeled by orthosteric ligands, arrestin, and a G protein mimetic. But whereas arrestin selects for pre-existing active-like substates, the G protein mimetic induces two new substates. Together, our study suggests i) a thus far unrecognized kinetic component to transducer selection, and ii) the available static structures do not represent the entire conformational ensemble observed in solution.
Publications
See Dr. Ziarek's publications on NCBI.
Contact
Contact Dr. Ziarek.
The Ziarek Lab is hiring highly motivated postdocs, graduate students and technicians.
Postdoctoral Fellow: Jin Gon Shim
Graduate Student: Tucker Shriver, III
Post-Baccalaureate Research Fellow: Kerem Kahraman
Research Data Analyst: Scott Robson